- Why does it say Windows can’t open this file? (New 9/14/04)
- Plotting variables in a data matrix against each other
- Isolating the cases associated with one group from a combined data set
- Problem related to CTL3D32.DLL
- Printing graphs
- How to use and get information about a macro such as distcomp() (corrected 9/23/04)
- How to enclose quoted things in macros
- How to format a file containing a matrix
- Problem in using downloaded macros
Why does it say Windows can’t open this file?
Q. I downloaded the new macros file mvgraphics.mac and attempted to open it in Windows XP.
I get a message stating:
Windows cannot open this file. To open this file, Windows needs to know what program created it.
You can select from the programs on your computer… .
If I then open MacAnova it just reinstalls MacAnova.
What am I doing wrong?
A. First off, the file you describe as being “MacAnova” is almost certainly not that, but is the installer file you downloaded and executed to install MacAnova. After I have used them, I either delete installer files or put them away out of sight. On my computer I have an Updaters and Installers folder. It can be handy to keep an installer if for some reason you need to repeat the installation.
The installer puts MacAnova and various help and macro files in a standard place. I'm not a Windows expert and I don't know exactly where that is. I do know it puts a shortcut to MacAnova for Windows on the XP start menu.
Secondly, there is probably no need for you to open macro files except from within MacAnova, so don't try to open a downloaded macro file by clicking on it. That is what led to the message stating that “Windows cannot open this file…’. Instead you should create a folder where you put downloaded MacAnova data and macro files and drag the macro file there. Or you can download it directly to that folder.
Then, before you use macros in the file do the following:
Cmd> addmacrofile("")
Find the file in the file navigation dialog box and select it. You will then be able to use macros in the file just like built-in functions.
Plotting Variables in a Data Matrix Against Each Other
Q. After I have downloaded the file T01_06 (Multiple sclerosis data), I want to display the two dimensional scatter plot of x2 and x4. MacAnova does not let me do that. I did the following and got that answer:
Cmd> plot(x2,x4) ERROR: argument 1 (x2) to plot() is not defined
How do I define x2 and x4? Or what else do I need to do? From reading the hand out on graphics, it is not clear to me.
A. First a word of clarification. T01_06 is not a data file but a data set or data matrix. It is one of the data sets in JWData5.txt.
There is a data file T1-6.DAT.DAT on the CD ROM that comes with the text but it is in a different format from the data file on JWData5.txt.
You can provide x2 and x4 to plot() in a couple of ways.
Cmd> data <- read("","t01_06") # find JWData5.txt T01_06 98 6 format ) Data from Table 1.6 p. 42 in ) Applied Mulivariate Statistical Analysis, 5th Edition ) by Richard A. Johnson and Dean W. Wichern, Prentice Hall, 2002 ) These data were edited from file T1-6.DAT on disk from book ) by making making column 1 be group number (1 or 2) and removing ) last column which was group coded as 0 or 1. ) ) Multple-Sclerosis data derived from evoked responses to visual ) stimuli recorded from the scalp of a subject. Two different visual ) stimuli S1 and S2 produced responses in the left (L) and right (R) ) eyes of subjects, some of whom had multiple sclerosis (MS). ) Col. 1: Group number, 1 = Non-MS group, 2 = MS group ) Col. 2: x1 = Age of subject ) Col. 3: x2 = S1L + S1R (Sum of left and right responses to stimulus S1) ) Col. 4: x3 = abs(S1L - S1R) ) Col. 5: x4 = S2L + S2R ) Col. 6: x5 = abs(S2L - S2R) Read from file "TP1:Stat5401:Data:JWData5.txt" Cmd> group <- data[,1]# group number Cmd> msdata <- data[group == 2, -1]#x1, x2, x3, x4, x5 for MS group Cmd> dim(msdata) # 29 cases and 5 variables (1) 29 5 Cmd> plot(msdata[,2],msdata[,4],xlab:"x2",ylab:"x4",\ title:"x4 = S2L+S2R vs x2 = S1L+S1R for MS subjects")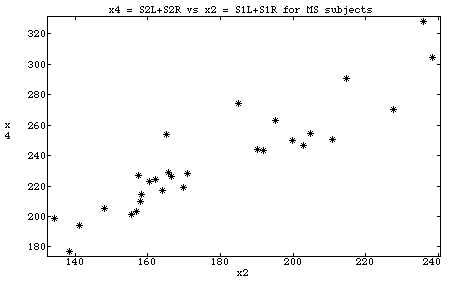
Here I am providing the two columns of the data to plot() using subscripts. msdata[,2] is all the rows and just column 2 of x and msdata[,4] is all the rows and just column 4 of msdata.
An alternative is to create separate MacAnova variables for each column of data. One easy way to do that is with makecols(). Here I create new variables x1, x2, x3, x4 and x5 from msdata:
Cmd> makecols(msdata,x1,x2,x3,x4,5) Column 1 saved as vector x1 Column 2 saved as vector x2 Column 3 saved as vector x3 Column 4 saved as vector x4 Column 5 saved as vector x5 Cmd> plot(x2,x4, xlab:"x2", ylab:"x4",\ title:"x4 = S2L+S2R vs x2 = S1L+S1R for MS subjects")
This is sort of a "black box" way to create separate variables. The "white box" way is
Cmd> x1 <- msdata[,1];x2 <- msdata[,2];x3 <- msdata[,3] Cmd> x4 <- msdata[,4];x5 <- msdata[,5] Cmd> plot(x2,x4 ......)
Here I have done several things things on one line, separating them by semi-colons.
The long and short of it is that, even if you have a data matrix named msdata, MacAnova doesn't know about x2 and x4, say, until you create them but does understand msdata[,2] and msdata[,4].
Isolating the cases associated with one group from a combined data set
Q. How can I isolate a subset of a data set for separate analysis?
A. Provided the subset is identifiable from the data set itself, this is relatively easy. For example, data set T01_06 in data file JWData5.txt contains data for both a multiple sclerosis (MS) group and a non-MS group. The group is coded 1 = non-MS and 2 = MS in column 1 of the data matrix. Here is how you might plot variable 4 (S2L + S2R) against variable 2 (S1L + S1R) for the MS group.
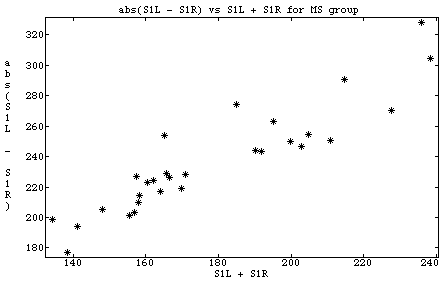
Cmd> data <- read("","t01_06", quiet:T) T01_06 98 6 format ) Data from Table 1.6 p. 42 in ) Applied Mulivariate Statistical Analysis, 5th Edition ) by Richard A. Johnson and Dean W. Wichern, Prentice Hall, 2002 ) These data were edited from file T1-6.DAT on disk from book ) by making making column 1 be group number (1 or 2) and removing ) last column which was group coded as 0 or 1. ) ) Multple-Sclerosis data derived from evoked responses to visual ) stimuli recorded from the scalp of a subject. Two different visual ) stimuli S1 and S2 produced responses in the left (L) and right (R) ) eyes of subjects, some of whom had multiple sclerosis (MS). ) Col. 1: Group number, 1 = Non-MS group, 2 = MS group ) Col. 2: x1 = Age of subject ) Col. 3: x2 = S1L + S1R (Sum of left and right responses to stimulus S1) ) Col. 4: x3 = abs(S1L - S1R) ) Col. 5: x4 = S2L + S2R ) Col. 6: x5 = abs(S2L - S2R) Read from file "TP1:Stat5401:Data:JWData5.txt" Cmd> group <- data[,1] # column 1 of data Cmd> nonms <- data[group == 1,-1]# take non ms rows, omitting col 1 Cmd> ms <- data[group == 2,-1]# take ms rows, omitting col 1 Cmd> list(nonms,ms) # find the dimensions of ms and nonms ms REAL 29 5 nonms REAL 69 5 Cmd> plot(ms[,2],ms[,4],title:"abs(S1L - S1R) vs S1L + S1R for MS group",\ xlab:"S1L + S1R",ylab:"abs(S1L - S1R)")
You could do this all in one step
Cmd> plot(data[group==2,3],data[group==2,5], ...)
but it is probably easier to include the intermediate steps. Doing it in one step, you select columns 3 and 5 instead of 2 and 4 because the whole data matrix includes the group number in column 1.
Several things to note:
- This data set has fairly extensive comments that describe its structure. In particular, the comments say that column 1 indicates whether a row is data for a nonMS or for a MS program. If you haven't seen them previously, you should always read the comments associated with a data set.
- LOGICAL subscripts are used to select rows. The expression group == 2 is a vector the same length as vector group (from column 1 of data) which has value T when group is 2 and F otherwise. When used as a subscript (data[group == 2, -1]), it selects all rows for which group is 2, that is all the MS rows.
- A negative subscript is used to indicate which columns are not to be selected. Thus data[group == 2, -1] selects all the MS rows, but omits column 1 which contains the MS/nonMS identifier. The result is you get the data for the MS group with out the group code.
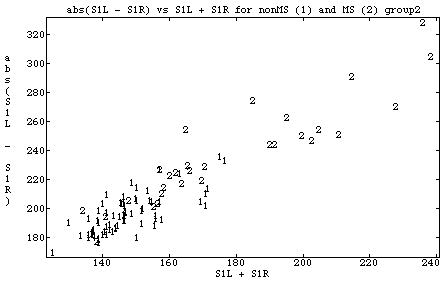
You might try plotting them both together, labeling non MS as 1 and MS as 2. This can be done as follows, using chplot().
Cmd> allcases <- data[,-1] Cmd> chplot(allcases[,2],allcases[,4],group,\ title:"abs(S1L - S1R) vs S1L + S1R for nonMS (1) and MS (2) group2",\ xlab:"S1L + S1R",ylab:"abs(S1L - S1R)")
The 3rd argument, group, is used by chplot() to label each point.
Back to topProblem related to CTL3D32.DLL
Q. I've downloaded MacAnova using the link on the class web site. The installation appears to be successful. However, when I try to run it I get the following message:
...CTL3D32.dll could not be found in the specified path c:\MACANOVA....
I've read all the installation instructions and gone through the process three times. It still doesn't work.
A. Here is a link to a page where you can download a version of CTL3D32.DLL for either Windows 95/98 or Windows NT (different files for 95/98 and for NT).
Link to ctl3d.htm
CTL3D3.DLL should be installed in your \WINDOWS\SYSTEM directory and not in the MacAnova directory. I have also read warnings you should not replace a newer version of CTL3D3.DLL with an older one.
Back to topHow can I print Graphs?
Q. When I select Print Graph on the File menu I get a message saying Printing is Disabled and giving instructions how to enable it. When I do enable printing and try to print again, MacAnova crashes. What am I doing wrong.
A. You are not doing anything wrong. The reason printing is disabled is that we have been unable to overcome the crashing problem. This doesn't happen in the Macintosh version.
The recommended way to print a graph is to paste it into a word processor document and then print the document. This is probably better in any event, because it allows you to add descriptive comments, and in some cases, get more than one graph per page.
Alternatively, you can limit yourself to low resolution plots, always using dumb:T in plotting commands. This causes graphs to be drawn using letters and other symbols directly in the command/output window.
Back to topHow to use and get information about a macro such as distcomp()
Q. I am not sure how to get distcomp() to work. I couldn't find anything about it in the MacAnova Users' Guide (though I may have missed it) or the Introduction to MacAnova. Could you give me a clue as to how to proceed with computing the generalized distances.
(9/23/04 The help() output has been corrected. In the MacAnova expression for d, (y - ybar') is not transposed when it’s on the left of %*% but is transposed on the right. Also the “guts” extracted from the macro have been updated.)
A. When x is a n by p data matrix,
Cmd> d <- distcomp(x)
computes a vector of length n = nrows(x) consisting of the generalized distances of each row of x from the sample mean.
Cmd> d13 <- distcomp(x[,vector(1,3)])
does the same, except only columns 1 and 3 of x are used.
You can find help on distcomp() by typing help(distcomp).
Cmd> help(distcomp)
Usage
distcomp(y) computes the generalized distances of each row of REAL
matrix y from the mean of all the rows. y must not have any missing
elements.
When y is n by p, the result is the length n vector
d = diag((y - ybar') %*% solve(s) %*% (y - ybar')'),
where ybar is the (column) vector containing the p column means and s is
the sample covariance matrix of y. The individual elements of d[i] are
d[i] = (y[i,]' - ybar)' %*% solve(s) %*% (y[i,]' - ybar),
Sometimes y = RESIDUALS, the matrix of residuals computed by manova().
Cross references
See also chiqqplot().
It may be instructive to take a look at what distcomp() actually does. Stripped of inessentials, here is the “guts” of what distcomp() does with a matrix x:
tmp <- tabs(x,covar:T,mean:T) # compute mean and variance matrix mean <- tmp$mean # p by 1 column vector y <- x' - mean # p by n transposed matrix of resids from mean s <- tmp$covar # p by p sample variance matrix d <- vector(sum(y * solve(s, y))) # value returned
The last line is a little tricky, first computing solve(s,y) = solve(s) %*% y, then multiplying each of its elements by the elements in y and then summing down columns (across variables for each case). This in fact computes
(x[i,]-mean') %*% solve(s) %*% (x[i,] - mean')'
for each i = 1, ..., n.
Back to topHow to enclose quoted things in macros
Q. I have been trying to automate a lot of tasks when doing homeworks. I tried to automate gamma Q-Q plots with the following macro:
Cmd> sqrtQQchi <- macro("chplot(sqrt(chiranks(ms)),\
sqrt(sort(distcomp(ms))),"o")")
ERROR: problem with input near
sqrtQQchi <- macro("chplot(sqrt(chiranks(ms)),sqrt(sort(distcomp(ms))),"o
It seems as though MacAnova reads the beginning quote for the character definition as the end quote for the macro command. How do I get it to process the whole text of the macro?
A. The problem is you are trying to include something quoted (namely "o") inside something that is already quoted. You can do it but have to take special precautions, namely to "escape" any internal quotes with a backslash so they won't be mistakenly taken to terminate an ongoing quote.
Here is your example corrected.
Cmd> sqrtQQchi <- \
macro("chplot(sqrt(chiranks(ms)),sqrt(sort(distcomp(ms))),\"o\")")
To quote from the help topic syntax,
The opening and closing double quotes are not part of the string. You include a double quote in a string by prefixing ("escaping") it with \ For example,
Cmd> a <- "He said, \"Hello\""assigns the string He said, "Hello" to variable a.
And help topic macro includes the following.
When you use macro() to define a macro, you should type any quotes '"' as '\"', as in the example. Similarly to include a backslash '\' you must type '\\'. In particular, quoted quotes in a macro must be typed as '\\\"' as inBack to topCmd> quotedecho <- macro("print(\"\\\"$0\\\"\")") Cmd> quotedecho(The answer is 42) "The answer is 42"
How to format a file containing a matrix
Q. Where can I find information on how to format matrix data so that it can be read into Macanova?
A. The simplest way to format matrix data is simply to put it in a text file, one row at a time. Then you can read it by
Cmd> x <- matrix(vecread("filename"),nc)' # note the transpose
where nc is the number of columns. For example, for the data for problem 1.4 on p. 39 of Applied Mulivariate Statistical Analysis, 5th Edition by Johnson and Wichern, the file with nc = 3 might look like the following.
126974 4224 173297 96933 3835 160893 86656 3510 83219 63438 3758 77734 55264 3939 128344 50976 1809 39080 39069 2946 38528 36156 359 51038 35209 2480 34715 32416 2413 25636
This works even when a row of the matrix is split between more than one line or there is more than one row per line. That is, it would work when the preceding data is formatted as
126974 4224 173297 96933 3835 160893 86656 3510 83219 . . . . . . .
or formatted as
126974 4224 173297 96933 3835 160893 86656 3510 83219 63438 3758 77734 55264 3939 128344 50976 1809 39080 . . . . . . . . . . . . . . . . . . . . . . .
You can get complete information on the format of files read by vecread() from help topic vecread_file.
You can get complete information on the format of files read by read() and matread() from help topic matread_file. Briefly, each matrix in the file is preceded by header lines of the form
matrixName nr nc ) 0 or more lines starting with ')' ) ....
where nr and nc are the numbers of rows and columns. After this line should come the data, one line per row. A file with the above data in this format might look like the following:
P01_04 10 3 ) Data from Problem 1.4 in Chapter 1 p. 39 in ) Applied Mulivariate Statistical Analysis, 5th Edition ) by Richard A. Johnson and Dean W. Wichern, Prentice Hall, 1998 126974 4224 173297 96933 3835 160893 86656 3510 83219 63438 3758 77734 55264 3939 128344 50976 1809 39080 39069 2946 38528 36156 359 51038 35209 2480 34715 32416 2413 25636
When a row is split among two or more lines you need a special line starting with ')' before the data. For example, when there are 5 numbers per line associated with a row, except possibly for the last line, the special line would be
)"%f %f %f %f %f"
where there are exactly 5 %f's. Every row of the matrix must start on a new line, and all lines for a row, except possibly the last, must have 5 data items. If there isn't such a header line like this one, there must be just one line per row. Here are the first few lines of a file containing the same data, with each case split between two lines:
Back to topP01_04 10 3 ) Data from Problem 1.4 in Chapter 1 p. 39 in ) Applied Mulivariate Statistical Analysis, 5th Edition ) by Richard A. Johnson and Dean W. Wichern, Prentice Hall, 1998 )"%f %f" 126974 4224 173297 96933 3835 160893 86656 3510 83219 . . . . . . . .
Problem in using downloaded macros
Q. I have not been able to use the correct macros for Hotelling's T2 posted on the Web page. I downloaded file hotelling.mac to the MacAnova folder. Here is what happens when I try to use one of the macros:
Cmd> t_sq <- hotell2val(x1,x2) WARNING: searching for unrecognized macro hotell2val near t_sq <- hotell2val( WARNING: macro hotell2val not found on external macro files ERROR: hotell2val is not function or macro near t_sq <- hotell2val(
What do I need to do?
A. What you are probably not doing is letting MacAnova know about file hotelling.mac.
When MacAnova looks for macros, it normally searches in the standard macro files. These include Arima.mac, Design.mac, Graphics.mac, MacAnova.mac, Math.mac, Mulvar.mac, Regress.mac and Tser.mac. You want to tell it to search in file hotelling.mac, too.
The list of files searched is in variable MACROFILES.
Cmd> MACROFILES (1) "Graphics.mac" (2) "Regress.mac" (3) "Design.mac" (4) "Tser.mac" (5) "Arima.mac" (6) "Mulvar.mac" (7) "Math.mac" (8) "MacAnova.mac"
Because hotelling.mac is not in the list, MacAnova can't find it.
You use addmacrofile() with getfilename() to add hotelling.mac to this list.
Cmd> addmacrofile(getfilename()) # find file hotelling.mac
At this point you find file hotelling.mac in the file navigation dialog box.
Cmd> MACROFILES (1) "hotelling.mac" (2) "Graphics.mac" (3) "Regress.mac" (4) "Design.mac" (5) "Tser.mac" (6) "Arima.mac" (7) "Mulvar.mac" (8) "Math.mac" (9) "MacAnova.mac" Cmd> t_sq <- hotell2val(x1, x2) WARNING: searching for unrecognized macro hotell2val near t_sq <- hotell2val(
Now the wanted macro is found.
The default use of addmacrofile() illustrated here, puts the file at the start of MACROFILES where it will be searched first. If you want to add a file at the end, to be searched last, use addmacrofile(fileName, T).
Cmd> addmacrofile("hotelling.mac",T)
Cmd> MACROFILES
(1) "Graphics.mac"
(2) "Regress.mac"
(3) "Design.mac"
(4) "Tser.mac"
(5) "Arima.mac"
(6) "Mulvar.mac"
(7) "Math.mac"
(8) "MacAnova.mac"
(9) "hotelling.mac"